| Project Description | Educational Outreach | Sponsors and Partners |
| Visit the alpha version of our new website in progress. |
|
The Challenge
How do the genetic makeup and environment interact to shape intricate developmental processes that lead to functional tissues, organs and organisms from undifferentiated cells? This has been a challenging question ever since biologists started wondering about development of multicellular organisms. The researchers have traditionally used microscopy, mutants and other methods to understand molecular and cellular bases for development. Recently, genomics is yet another tool added to the developmental biologists' arsenal. With the advances in biological knowledge, imaging instrumentation, applied biomathematics, and computing, it is now becoming possible create and apply computational modeling to integrate multidisciplinary approaches and different types of biological data in studying development.
The Project
Educational Outreach
The researchers also plan to develop, evaluate, and introduce a new set of techniques for high school and pre-service science teachers as well as to undergraduate students. Outreach activities will culminate in a summer institute in which 30 high school students will develop a public kiosk to display the "silicon plant" model for exhibit at the Huntington Botanical Gardens, which hosts 500,000 visitors per year. This program holds remarkable promise for linking cutting-edge knowledge and techniques with K-12 teachers' and students' understanding of plant development and integrative biology. Go to top of Page
Sponsors and Partners
 Sponsored by the National Science Foundation Frontiers in Integrative Biological Research (FIBR) Program     
|
 The sun never sets on The Computable Plant! The sun never sets on The Computable Plant! 
|
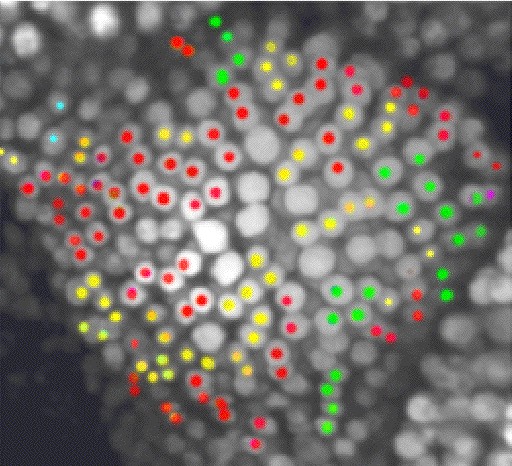 Biological experimentation in this project uses cleverly designed transgenic plants with marker proteins and modification of known developmental genes. The bioinformatics component will manage a vast amount of image and knowledge data. The mathematical component will automatically generate specialized, efficient simulation code from models and link it to suitable bioinformatic datasets through pattern recognition, machine learning, and regulatory circuit inference algorithms. Extensive visualization, image processing, and optimization software will fit these predictive models to image data. Scientific objectives of this effort include the development and use of such mathematical modeling software for plant development, as well as its use to explore alternative hypotheses in silico and to guide in vivo experiments. Thus the project will involve a working loop from experiments, through bioinformatics and mathematical modeling, and back to experiments.
Biological experimentation in this project uses cleverly designed transgenic plants with marker proteins and modification of known developmental genes. The bioinformatics component will manage a vast amount of image and knowledge data. The mathematical component will automatically generate specialized, efficient simulation code from models and link it to suitable bioinformatic datasets through pattern recognition, machine learning, and regulatory circuit inference algorithms. Extensive visualization, image processing, and optimization software will fit these predictive models to image data. Scientific objectives of this effort include the development and use of such mathematical modeling software for plant development, as well as its use to explore alternative hypotheses in silico and to guide in vivo experiments. Thus the project will involve a working loop from experiments, through bioinformatics and mathematical modeling, and back to experiments.