The Computable Plant
Eric Mjolsness, a computer scientist at University of California, Irvine and Elliot Meyerowitz, a plant developmental biologist at California Institute of Technology will work together to provide a quantitative and cellular description of plant development. They will study meristem development in Arabidopsis thaliana, the model plant that has been used extensively in contemporary plant biology research. Meristems are the inner plant tissues, where regulated cell division, pattern formation and differentiation give rise to plant parts like leaves and flowers. The investigators at Caltech will use green fluorescent proteins to mark specific cell types in the apical meristem and image their lineages through meristem development and differentiation leading to specifi arrangement of leaves and reproductive growth. Automation of image acquisition and analysis will help them generate and visualize a vast amount of data, which will be used by the UCI investigators to model cells and their patterns in the developing meristem and simulate developmental processes under different conditions. These simulations will result in predictions that will be tested experimentally using mutants, altered hormone gradients, and other manipulations.
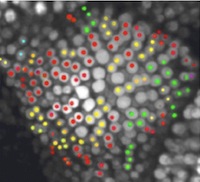
Biological experimentation in this project uses cleverly designed transgenic plants with marker proteins and modification of known developmental genes. The bioinformatics component will manage a vast amount of image and knowledge data. The mathematical component will automatically generate specialized, efficient simulation code from models and link it to suitable bioinformatic datasets through pattern recognition, machine learning, and regulatory circuit inference algorithms. Extensive visualization, image processing, and optimization software will fit these predictive models to image data. Scientific objectives of this effort include the development and use of such mathematical modeling software for plant development, as well as its use to explore alternative hypotheses in silico and to guide in vivo experiments. Thus the project will involve a working loop from experiments, through bioinformatics and mathematical modeling, and back to experiments.