SigMech
kMech
[Yang 2005] is an enzyme mechanism modeling tool designed for the
mathematical modeling of enzymes. It comprises a collection of
single and multiple substrate enzyme reactions. Over the years
the library of requested enzymatic reactions implemented in the kMech
utility and correspondingly in Sigmoid has grown substantially.
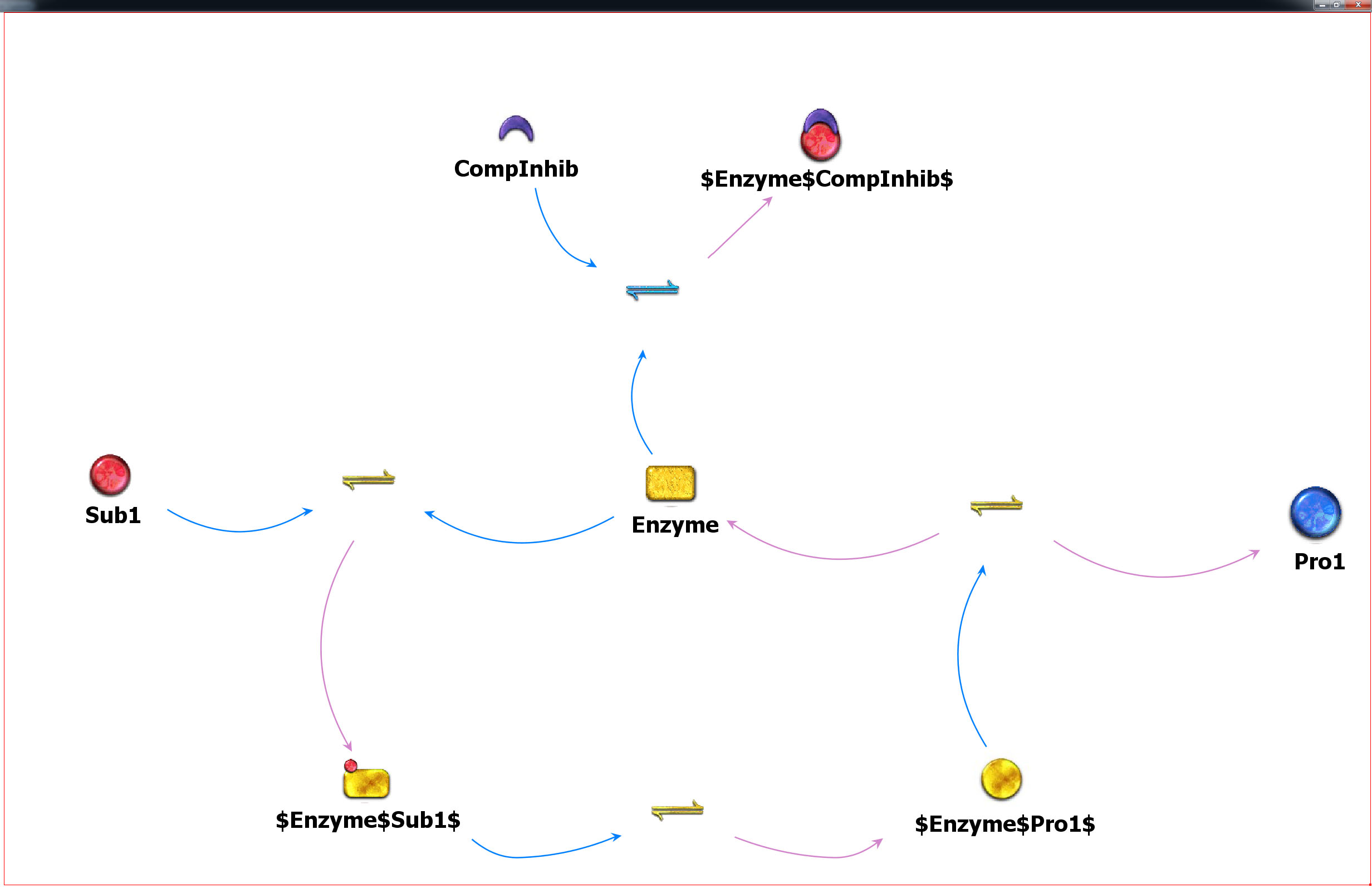
A new generalized version of the kMech enzyme mechanism modeling tool has been developed. With this utility, SigMech, the approximately 35 existing enzyme mechanism models expressed explicitly in the kMech/Sigmoid platform can be expressed implicitly by a single parameterized input notation. Subsequent sub-reactions can be generated procedurally, and any potential “new” kMech enzyme mechanisms that fall within the pattern abstracted from previous motifs need not be created explicitly.
The highly parameterized reaction types of SigMech, together with the kMechReaction classes in the Sigmoid schema, provide a parameterized representation that encompasses all previous Compound reactions that were present in the Sigmoid schema, and can generate these and other novel reaction mechanisms from the cross product of the valid SigMech input parameters. This development effectively renders kMech integration a solved problem within the scope of previous reaction motifs, and will vastly reduce the need for frequent updates of the Sigmoid schema, database, middleware translation and GUI.
SigMech combines the reaction motifs of previous kMech reaction mechanisms, with the conversion phase that was implemented in the three-stage xCellerator reaction (ThreeStageCatalytic in the Sigmoid schema) yielding a greater parameter space of possible enzymatic reaction mechanisms that can be generated on the fly inside SME, and that can consequently be simulated by Mathematica/ xCellerator.
SigMech integrated with the Sigmoid Model Explorer can be found here as a Java .jar executable.